Metabolic Pathway Design Technology
Tomokazu SHIRAI
Deputy Team Leader
Cell Factory Research Team, Center for Sustainable Resources Science,
Institute of Physical and Chemical Research (RIKEN)


Tomokazu SHIRAI
Deputy Team Leader
Cell Factory Research Team, Center for Sustainable Resources Science,
Institute of Physical and Chemical Research (RIKEN)


It enables designing artificial metabolic reaction pathways, and further, designing intracellular metabolism for achieving high productivity to produce target compounds. In addition, based on the blueprints, candidate genetic modifications (deletions and enhancements) can be extracted and proposed.
Example of ApplicationUseful aromatic compoundsω-3 polyunsaturated fatty acid-containing lipids
In the biotechnology industry, many useful compounds have been conventionally produced by utilizing microbial "fermentation." In recent years, the technology for producing useful compounds by utilizing microbial "fermentation" is used as a technology for utilizing non-fossil raw materials. This technology, where sugars, etc. that are produced through carbon dioxide fixation by plants and photosynthetic microorganisms are utilized as a carbon source, is used to have genetically modified microorganisms produce target useful compounds. Europe and the United States are leading the way in microbial production technology using this "synthetic biology," and cataloging of the production technologies using fermentative production is already in progress for many general-purpose chemicals. Concrete examples of the general-purpose chemicals that have been cataloged are general-purpose polymer raw materials, such as bioethanol, which replaces fossil fuels in automobiles, lactic acid, which is a raw material for polylactic acid, 1,3-propanediol, γ-aminobutyric acid and 4-aminocinnamic acid. Furthermore, as an example of manufacturing industrial application, a US bio-venture Genomatica and BASF (Germany) have succeeded in jointly producing more than 50,000 tons per year of 1,4-butanediol (BDO), which is a core general-purpose compound.
For the microbial production of useful compounds, it is essential to have a technology that optimally designs "metabolism" including not only the intracellular carbon flow but also the balances between energy production and consumption and between oxidation and reduction. This is because such a technology allows the understanding of the intracellular phenotype and the information obtained can be applied to the metabolism design of the target cell and to subsequent breeding. However, there is a limit to the human brain's capacity to think about more than 1,000 metabolic reactions occurring in a cell. Therefore, the computing power of computers is indispensable. Especially in recent years, due to the acceleration of the annotation process by the innovations in the genome sequencing technology and information processing technology, it has become possible to describe all metabolic reactions on a computer at the genome scale level. In other words, a technology for predicting the metabolic behavior of microbial cells in a certain environment has been established (genome scale model: GSM). Currently, research is being actively conducted by systematically performing the process from cell metabolism design using GSMs to verification by actual experiments, in order to improve the target compound productivity in a high-throughput manner. However, the existing GSMs cannot predict or design biosynthetic pathways for non-natural compounds. In addition, design using metabolic reactions that occur in non-host cells is difficult. We are developing 2 metabolic pathway design tools that can be a technology to solve these problems.
An overview of this tool is provided below (Fig. 1).
1. From the databases containing metabolic and enzyme reactions, such as KEGG (http://www.genome.jp/kegg/) and BRENDA (http://www.brenda-enzymes.org/), the concept of individual enzymes was uncoupled, and only the chemical reaction patterns were described. Then, similar chemical reaction patterns were reclassified as one chemical reaction and the computer was made to learn the reclassified patterns (Fig. 1 (a)).
2. In the learning process, the precursor and product in each reaction were described using a notation called SMILES, and then the reaction mechanism was described entirely using a notation called SMIRKS (Fig. 1 (b)).
3. In the actual simulation, the target compound is described using the SMILES, and the description is used as input data. Then, based on this, the precursors are retro-synthesized randomly and comprehensively. The simulation is successful if a compound known to exist in vivo is present in the retro-synthesized precursors. In other words, if the designed artificial metabolic reaction takes place using the known biological compound as a starting material, the target compound can be biosynthesized (Fig. 1 (c)).
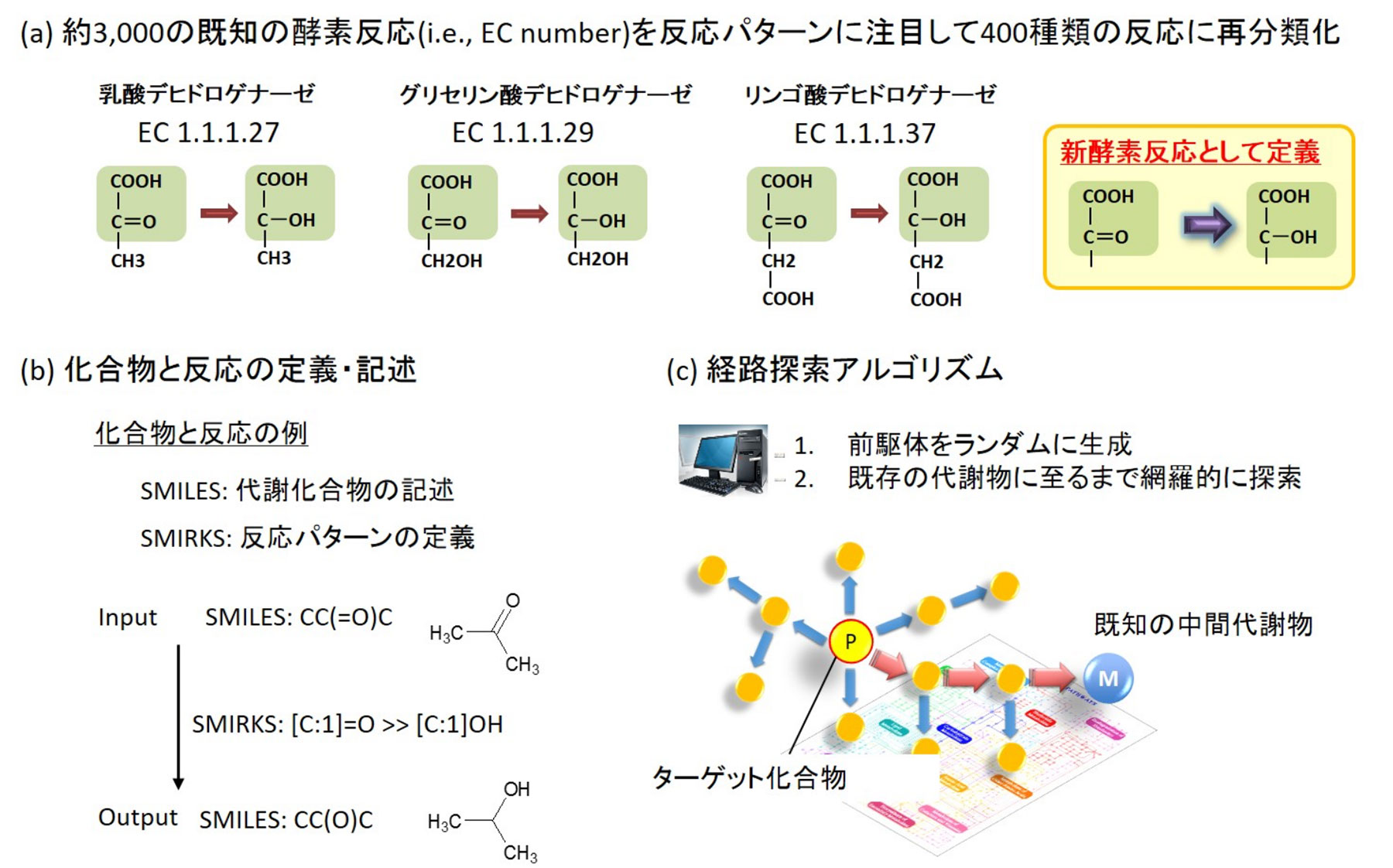
図1.人工代謝反応を設計するBioProVの概要
M-Path1 developed by Michihiro Araki, Professor, at Kyoto University has a different algorithm but the same concept, and is known as a tool for designing artificial metabolic pathways.
The existing GSMs allow metabolic pathway design only within the scope of metabolic reactions that occur in the host cell. Therefore, we have developed a tool that enables hybrid metabolic pathway design in order to efficiently produce target compounds, by comprehensively adding metabolic reactions that occur in organisms other than host cells.2 The outline of this tool is as follows.
1. From all species' metabolic reactions contained in the KEGG database (http://www.genome.jp/kegg/), metabolic reactions that occur in the host cell to be used were excluded, and the remaining reactions were stored in the database.
2. From the database prepared, the reaction pathways that connect to the GSMs of the host cell were selected to construct a hybrid metabolic pathway (HyMeP) (Fig. 2).
3. Using the constructed HyMeP, we have designed an efficient metabolic pathway that can achieve the maximum production of target compounds.
Drawing an ideal metabolic pathway blueprint using the HyMeP enables rapid and rational metabolic pathway design.
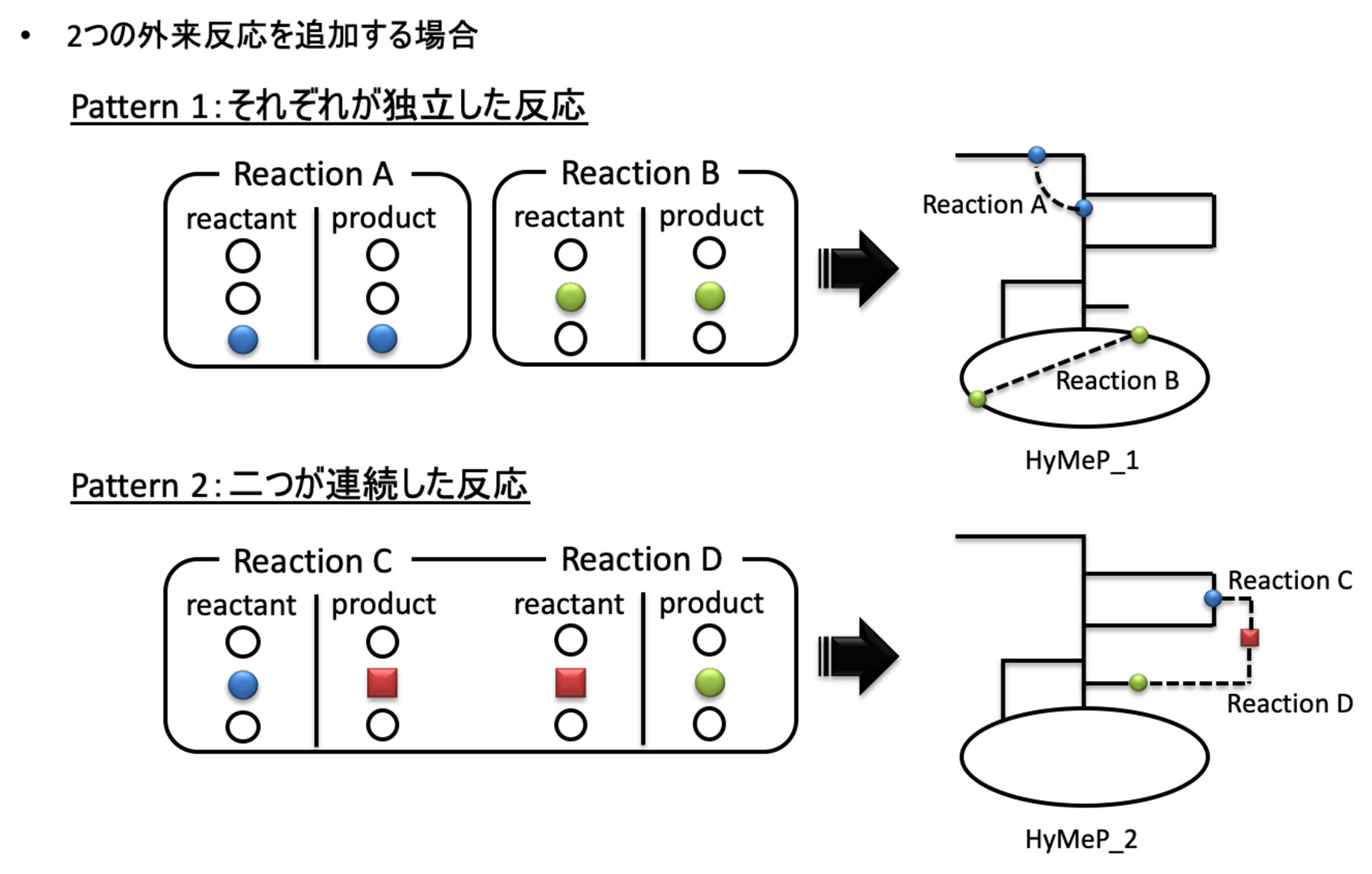
図2.2つの外来反応を宿主の代謝モデルに付加しHyMePを構築するスキーム
In order to efficiently produce useful compounds using microbial cells, it is indispensable to design an optimal intracellular metabolic pathway by computation. We have developed a tool BioProV, which proposes new metabolic pathways, and a tool HyMeP, which enables the efficient production of useful compounds by hybrid metabolic pathway design. By combining these 2 tools, an optimal metabolic pathway can be designed, for example, to have microbial cells produce petroleum-derived non-natural useful compounds. In the future, it will be important to highly activate the metabolic reaction that takes place in the designed metabolic pathway.
Last updated:December 25, 2023