Long-chain DNA Synthesis Technology
Kenji TSUGE
Specially-Appointed Associate Professor
Graduate School of Science,
Technology and Innovation, Kobe University


Kenji TSUGE
Specially-Appointed Associate Professor
Graduate School of Science,
Technology and Innovation, Kobe University


It enables accurate construction of designed long-chain DNA in a short period of time at a low cost.
To accelerate the development of smart cells, how fast the DBTL cycle can be run is the key. Looking at each step, the Build step, where bacterial strains with sequences designed at the Design step are actually created, takes the longest time. Within the step, the synthesis of long-chain DNA with designed sequences has required a long period of time, specifically several months.
This technology uses the ordered gene assembly in Bacillus subtilis (OGAB) method, which we developed to construct long-chain DNA (Fig. 1).1,2 The OGAB method is a method for assembling multiple DNA fragments using the plasmid transformation system of B. subtilis. Utilizing the specificity of 3 to 4 bases at the end of a DNA fragment, the method enables one-step assembly of a maximum of more than 50 DNA fragments. In order to construct long-chain DNA with arbitrary sequence, it is necessary to prepare a double-strand DNA fragment having a size of several hundreds to several thousands of base pairs (bp), as required by the OGAB method, using chemically synthesized single-strand DNA as a starting material. So far, this synthesis has been outsourced to a contract DNA synthesis company. When ordering the synthesis of 50 DNA fragments at a time, however, cases such as that where the synthesis of some fragments is rejected or where the synthesis is attempted but fails and is eventually rejected occur almost invariably although many DNA fragments are successfully synthesized. In such cases, the synthesis is outsourced once again to another contract DNA synthesis company or the design of the DNA fragments is modified, and sometimes it takes as long as 2 months to finally prepare all the necessary DNA fragments.
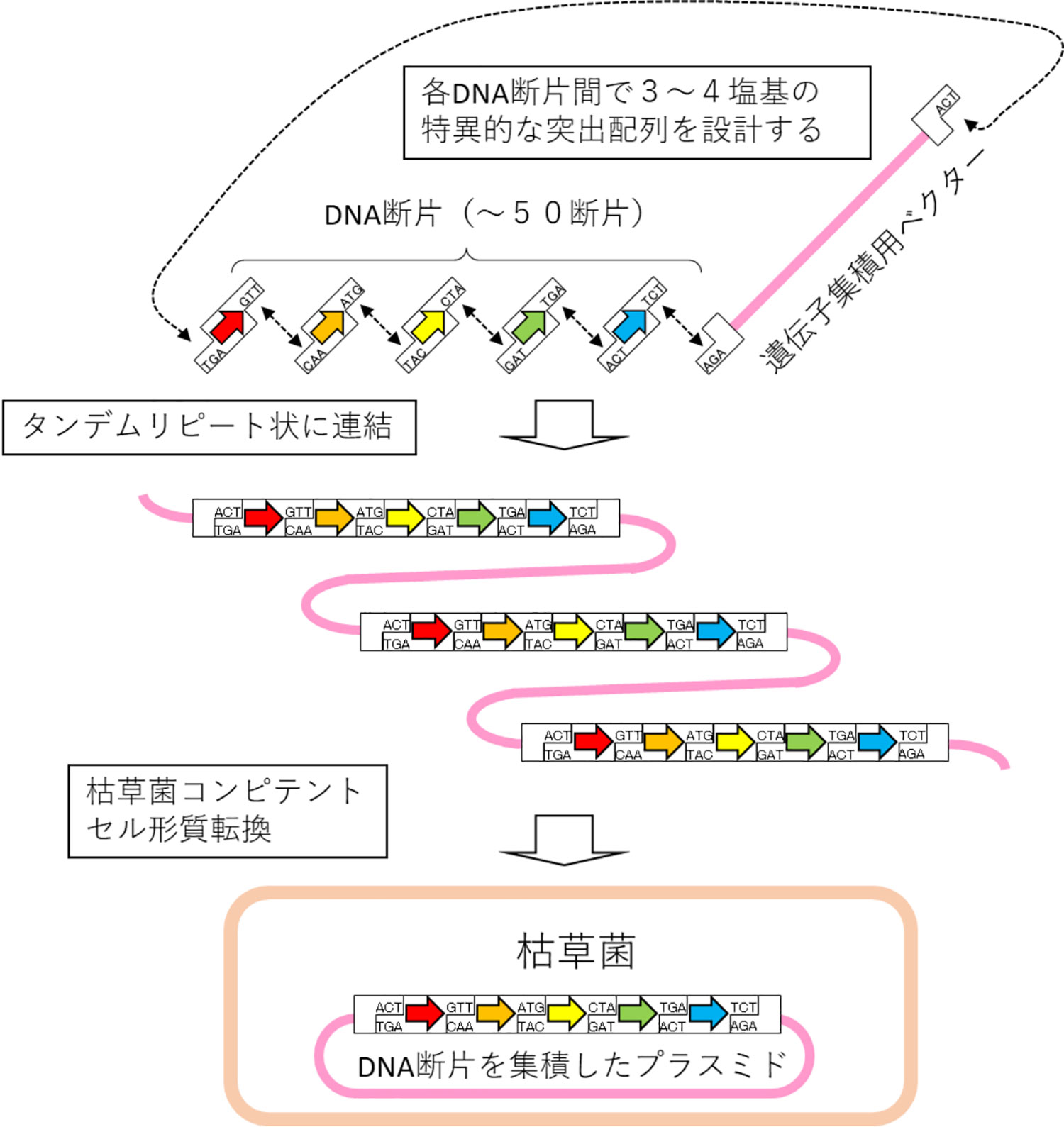
図1.OGAB法による遺伝子集積の概要
Therefore, in order to shorten the time taken for DNA fragment preparation, a total system capable of mass preparation of long-chain DNA constructed through chemical synthesis was established in Kobe University, and technologies were developed from a comprehensive perspective (Fig. 2). Individually, through joint research with Nihon Techno Service Co., Ltd., we have developed a DNA chemical synthesizer specializing in the synthesis of long-chain DNA, which synthesizes 96 single-stranded DNAs of 200 bases in about 1 day at a low cost (Fig. 2, left). In addition, we have also developed a new PCR method for annealing and elongating chemically synthesized DNA using DNA complementarity. This made it possible to prepare double-stranded DNAs of any sequence in about 3 days. In addition, the process in which this double-stranded DNA fragment is cloned in E. coli and only the selected clones having the correct base sequence are used as a material in the OGAB method was substantially automated by a liquid dispensing robot (Fig. 2, center). As a result of these research and development activities, it has become possible to produce long-chain DNA of about 30 kb in a short period of about 2 weeks at a cost of several yen per base. Thus, we have achieved a significant reduction in the time and financial costs required by conventional methods.
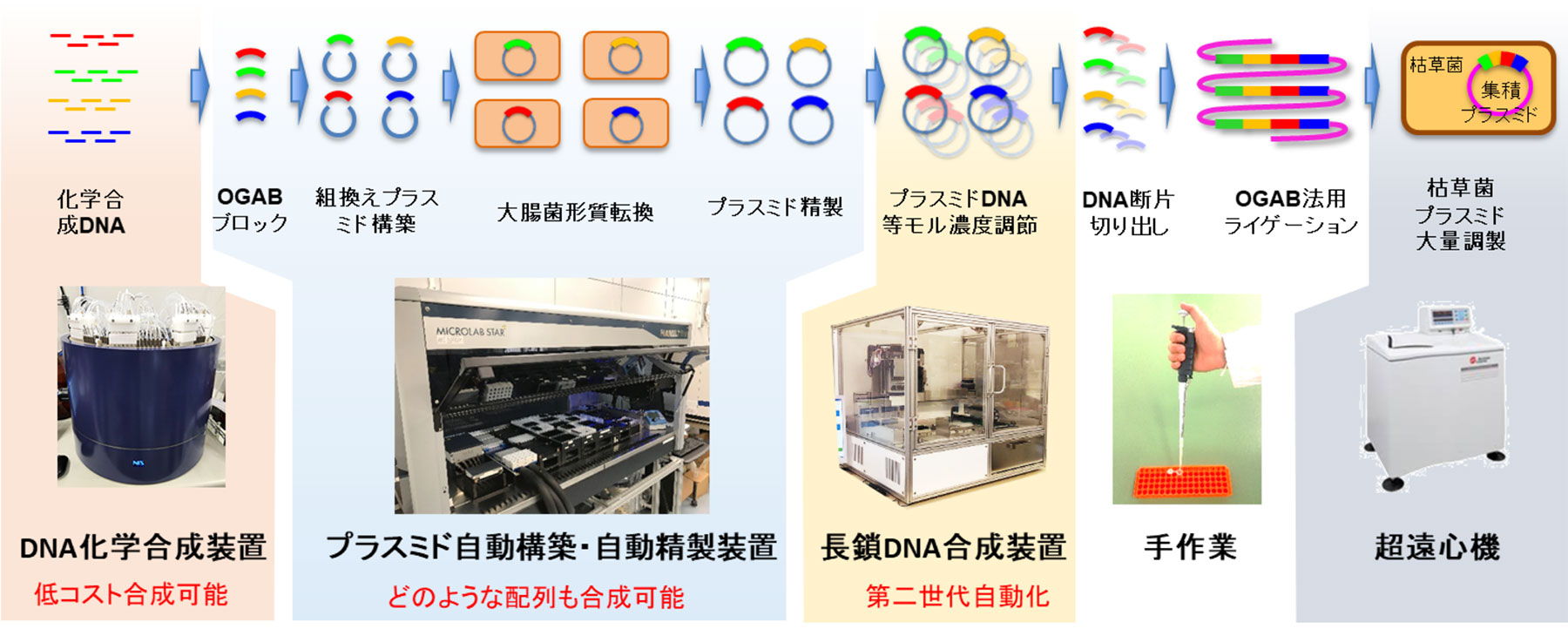
図2.神戸大学に構築した長鎖DNA合成トータルシステム
Japanese Patent No. 4479199 "Method for producing plasmid containing insertion DNA unit"
Japanese Patent No. 6440636 "Method for preparing DNA unit composition and method for creating DNA assemblies"
Last updated:December 25, 2023