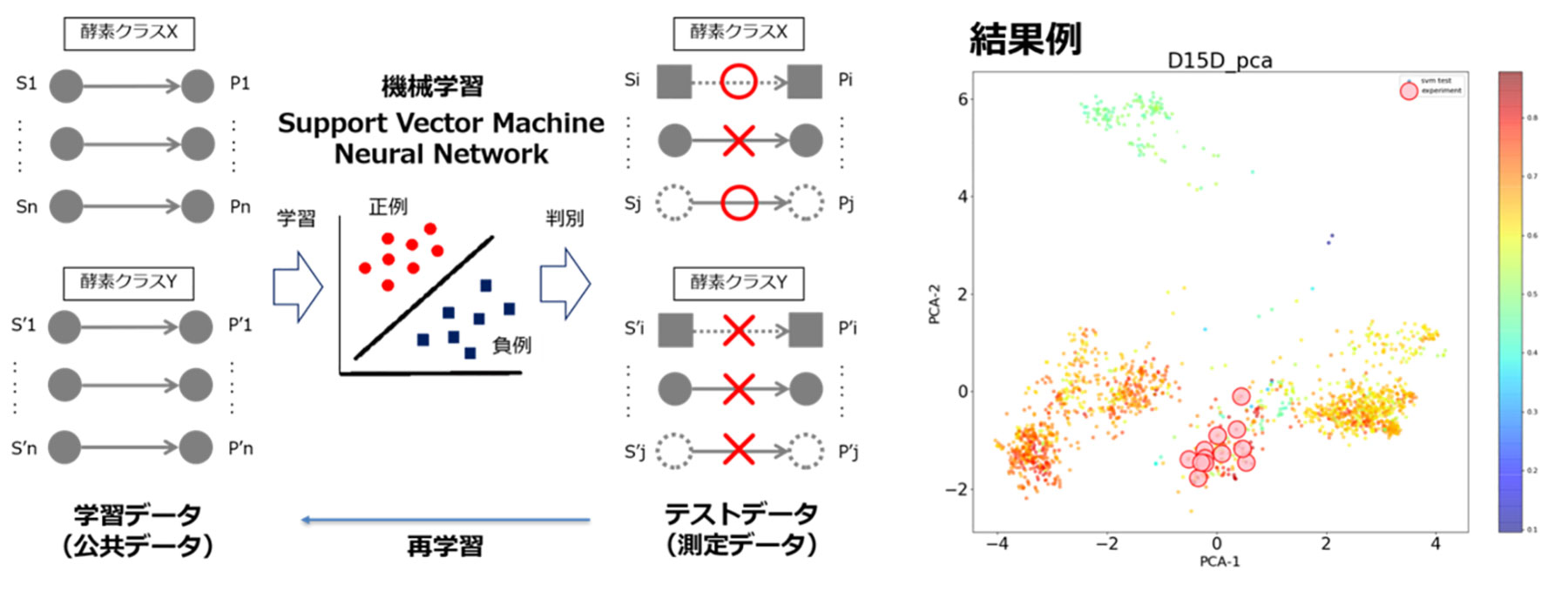
Estimated multiple enzyme gene candidates, either unknown or known, will appear in metabolic pathways constructed by metabolism design. Therefore, the selection of enzyme genes in the actual construction of a metabolic pathway is an important issue. However, at present, also regarding this point, each researcher manually searches the information separately contained in various enzyme reaction databases such as KEGG and BRENDA, as well as literature and patent information, and makes decisions based on his or her intuition. Under these circumstances, there is a strong demand for the development of an efficient and reliable method for selecting enzyme genes.
The machine learning method is an approach to discriminate and classify test data to be predicted, based on the learned data. It has been applied in various fields, due to an increase in data volume and an improvement in computing power in recent years. This field is no exception; if the machine learning method enables learning based on the known enzyme reaction data and leads to the discovery of new enzyme reactions, it can be a very useful method. We aim to extract features that cannot be identified by conventional sequence comparison methods and clustering methods, by developing a machine learning method that considers the combination of substrates/products and enzyme amino acid sequences and a deep learning-based method for extracting the features of an enzyme's amino acid sequences.
Strengths in the Industrial Arena
This technology can contribute to the discovery of new useful enzyme genes, in the search for enzyme genes to realize new metabolic pathways and the search for enzyme gene variants having new substrate specificities and activities in the known metabolic pathways.