Expression Regulation Network Construction Technology
Sachiyo ABURATANI
National Institute of Advanced Industrial Science and Technology (AIST)


Sachiyo ABURATANI
National Institute of Advanced Industrial Science and Technology (AIST)


We aim to understand the underlying mechanisms of phenomena occurring in microbial cells during material production to regulate the phenomena as one operation system. For that purpose, we propose candidate genes to be modified for artificial regulation, by selecting genes contributing to material production from gene expression data and expressing their interactions as a network model.
In order to achieve efficient material production using microorganisms, it is necessary to understand the underlying mechanisms of phenomena occurring in microbial cells during material production and regulate the phenomena as an operation system. For understanding and utilizing this complex "system" in living cells, a network model of gene expression regulation is useful. By constructing the regulatory relationship between genes as a network model, we were able to express the process occurring in living cells in a causal graph. This has led us to the development of a technology that allows the search for modification sites necessary for the bottleneck search in the process and for promoting efficiency.
As a technology for estimating gene groups that contribute to material productivity, we have developed a work flow that allows the gene groups related to material production to be extracted with higher accuracy, by combining statistical testing, correlation analysis and logical operations.
After selecting candidate genes as parts of the regulatory system, network modeling of the regulatory relationships between genes was performed. Using the network model, we have identified the genes which can be regulatory factors to control the production level of a target material in the material production process, from the selected genes groups that are likely to correlate with the production levels of target materials. In this study, we are working on the high-accuracy estimation of network structures, by combining Bayesian networks1 and the structure equation modeling2,3 developed by the National Institute of Advanced Industrial Science and Technology (AIST).
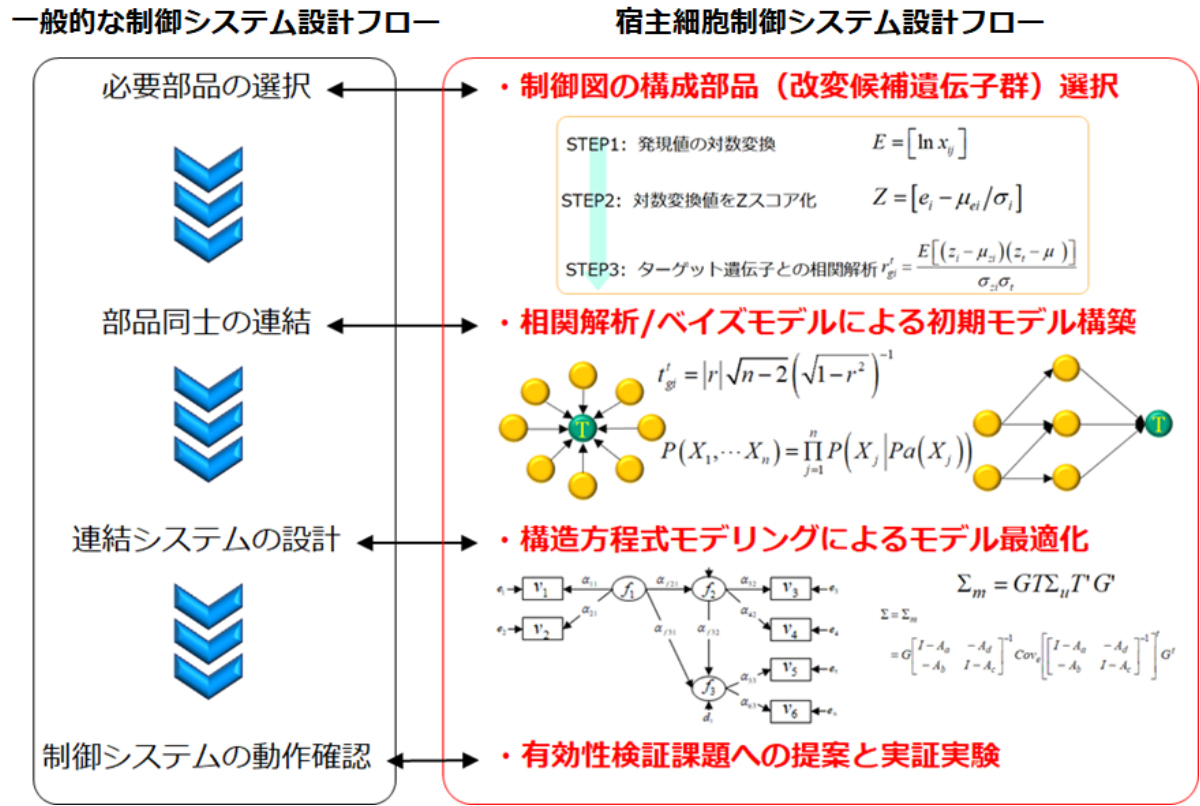
So far, we have proposed many candidate genes to be modified that could not be searched for by the conventional breeding approach, and succeeded in about a 2- to 10-fold increase in the production level. Thus, this technology can contribute to the creation of highly productive microorganisms. Furthermore, this technology has a wide scope of application: it is applicable not only to gene expression data, but also to any numerical data. It is a technology that can be applied not only to material production using microorganisms, but also to cases where the regulation of certain types of systems that are "black boxes" is required.
1) N. Friedman et al. : J. Comput. Biol., 7(3-4), 601-620 (2000)
2) S. Aburatani : Gene Reg. Sys. Biol., 5, 75-88 (2011)
3) S. Aburatani and H. Toh : Encyclopedia of Information Science and Technology, 3rd Edition, pp.458-478, IGI Global (2015)
Last updated:December 25, 2023