Transgene Sequence Design Technology
Tomoshi KAMEDA
National Institute of Advanced Industrial Science and Technology (AIST)


Tomoshi KAMEDA
National Institute of Advanced Industrial Science and Technology (AIST)


This technology enables controlling (increasing or decreasing) the expression levels of microbially-produced proteins by using a gene sequence modification method focusing on the mRNA secondary structure and the frequency of codon usage.
In this research, we first analyzed information on protein production experimental data on Rhodococcus sp. owned by the National Institute of Advanced Industrial Science and Technology (AIST). The data contained information on the gene sequences of 204 genes, with a link to their protein production levels. More specifically, by calculating various sequence feature values from the genetic sequences, the correlation between protein expression levels and sequence feature values was evaluated.
As a result, the stability of the mRNA secondary structure at the head of the sequence and the frequency of codon usage (Codon Adaptation Index: CAI) were highly correlated with protein production levels.
sequences to prevent the formation of secondary structures and also to have high CAI values.
We have confirmed that this method can improve protein expression levels with a high probability, the success rate being as high as 75%.
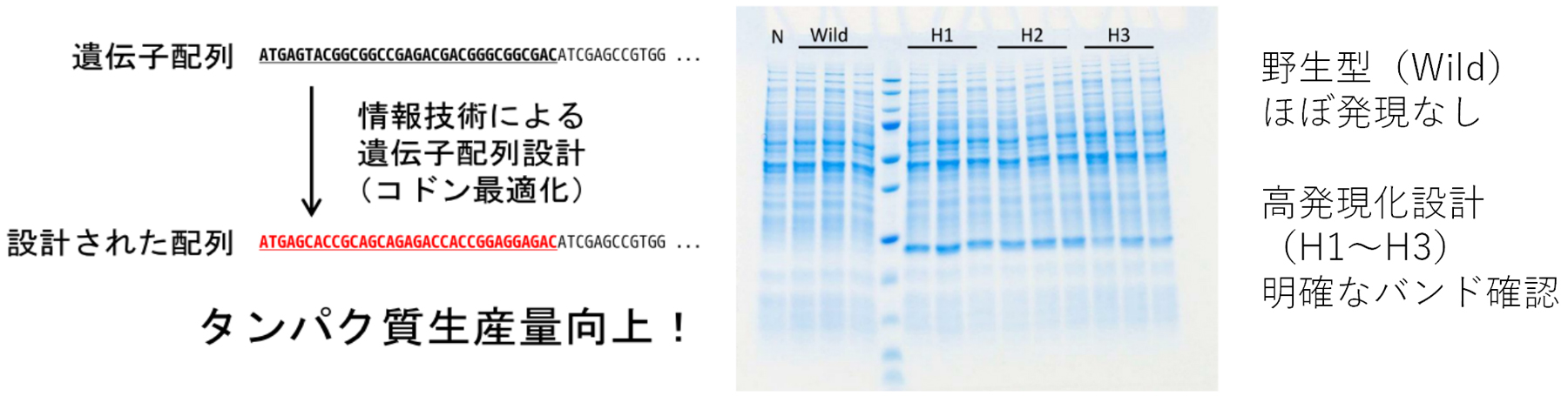
Although the effectiveness of this method was verified in Rhodococcus sp.,1 this method can be applied to material production in various microbial hosts other than actinomycetes. We have actually confirmed its effectiveness in Escherichia coli, Corynebacteria, etc.
In addition, we have succeeded in increasing the expression of target materials that are difficult to be expressed, such as membrane proteins.
Since this method modifies the sequence at only the 5'-terminal head region of a gene sequence, it also allows inexpensive synthesis.
1) Y. Saito et al.: Sci. Rep., 9(1), 8338 (2019)
Last updated:December 25, 2023