High production of alkaloids in Escherichia coli based on metabolic pathway design, enzyme design and metabolomic analysis


Metabolic Pathway Design, Knowledge Extraction and Learning from Literature, etc.(Proposing Enzymes Utilizing Machine Learning and Knowledge Base Supporting Smart Cell Designing), High-accuracy Metabolomic Analysis
Kobe University, Ishikawa Prefectural University
To produce benzylisoquinoline alkaloids (BIAs), which are plant-derived natural products, at high levels utilizing microorganisms
BIAs, which are plant-derived natural products, contain many important compounds as raw materials for pharmaceuticals, including opioid analgesics. Conventionally, commercial products have been obtained by extraction from plants, but there were problems in terms of efficiency and cost. Although studies on Escherichia coli- and yeast-based production have been reported in recent years, there has been a demand to improve productivity for practical application due to their low production levels. Therefore, we aim to resolve the bottleneck of BIA production, by using the Design-Build-Test-Learn (DBTL) workflow, which combines bioinformatics and synthetic biology.
Previous studies have shown that the activity of an enzyme intracellularly producing tetrahydropapaveroline (THP), a BIA precursor, is weak. Therefore, the resolution of this bottleneck has been a critical issue. In order to create a smart cell that produces highly functional and useful materials in large quantities, it is necessary to design a metabolic pathway that increases the production level and yield, and to transduce genes realizing the designed structure into host microbial cells.
For this purpose, using "M-path," a bioinformatics-based metabolic pathway design tool developed by Dr. Araki, we have designed a new metabolic pathway capable of shortcutting the conventional metabolic pathway bottleneck to contribute to an improvement in the BIA productivity. In addition, we have also succeeded in engineering an enzyme that not only has the new pathway, but also the conventional pathway in a well-balanced manner, by discovering enzymes that constitute the new shortcut pathway from nature and modifying the amino acid sequence through the structure simulation. A verification study conducted by transducing genes related to the designed metabolic pathway and enzymes into Escherichia coli has revealed that both metabolic pathways function well within the bacterium and successfully increased the production level of THP, a metabolic intermediate in the BIA biosynthesis, more than twice the conventional level. Moreover, by metabolomic analysis of the producing bacterium, we have found a metabolism rule that leads to further improvement in productivity. We examined the culture conditions based on this rule, and succeeded in increasing the production level of reticuline, a BIA, more than 7-fold the conventional level. Thus, we are one step closer to the realization of BIA production by the microbial fermentation method.
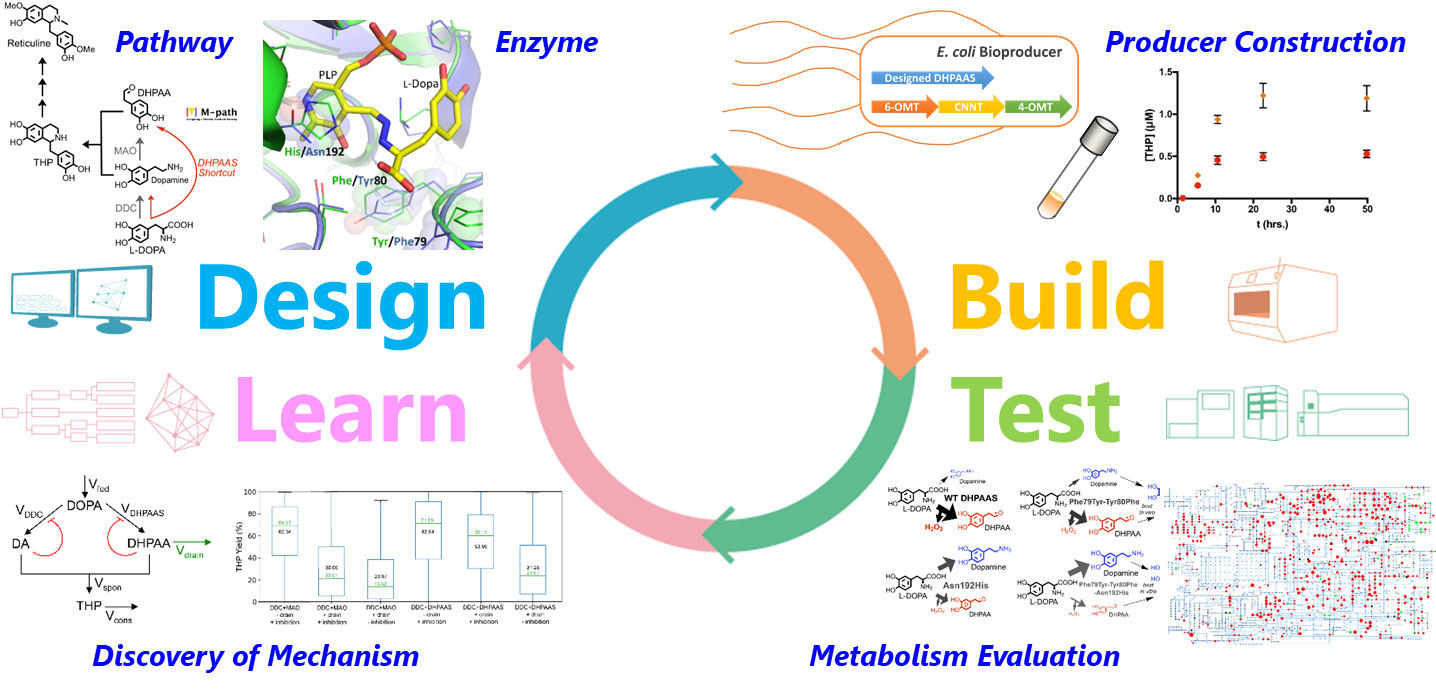
Vavricka, CJ., Yoshida, T., Kuriya, Y., Takahashi, S., Ogawa, T., Ono, F., Agari, K., Kiyota, H., Li, J., Ishii, J., Tsuge, K., Minami, H., Araki, M.*, Hasunuma, T.*, Kondo, A.: Mechanism-based tuning of insect 3,4-dihydroxyphenylacetaldehyde synthase for synthetic bioproduction of benzylisoquinoline alkaloids, Nature Communications, 10, 2015(2019)
Last updated:December 25, 2023